Difference between revisions of "Gene Editing"
(added more history) |
(added TALENs section) |
||
| Line 8: | Line 8: | ||
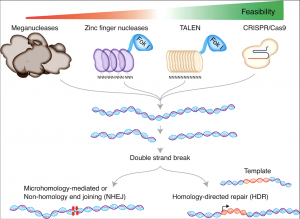
In 1985, possibly the first major technique, for highly targeted genome editing, was the discovery of zinc finger nucleases (ZFN), which improved the effectiveness of gene targeting. ZFNs help “recognize” DNA, which makes it possible to design ZFNs for a variety of genomic targets varying throughout different cells and organisms. <ref> [https://www.synthego.com/learn/genome-engineering-history “Full Stack Genome Engineering.”] Synthego, </ref> ZFNs are fusion proteins composed of DNA-binding domains that recognize and bind to a specific set of three base pairs. They can be combined to recognize longer DNA sequences. For example, specificity to a nine-base-pair target sequence would require three ZFN domains fused in tandem. <ref> Fridovich-Keil, Judith L. [https://www.britannica.com/science/science "Gene Editing"] Encyclopædia Britannica, Inc., 20 Feb. 2020 </ref> Researchers then fused the DNA cleaving (cutting) enzyme Fok1 to the ZFN binding domain to essentially create “genomic scissors” which can cleave DNA at a specified location, creating a double stranded break (DSB) in the DNA. <ref> [https://en.wikipedia.org/wiki/Zinc_finger_nuclease “Zinc Finger Nuclease"] Wikipedia, Wikimedia Foundation, 29 Jan. 2020, </ref> A repair template is then introduced, so a fraction of the cells will undergo [https://en.wikipedia.org/wiki/Homologous_recombination homologous recombination], where the cell incorporates the gene of interest into its genome at the specific point. <ref> [https://www.synthego.com/learn/genome-engineering-history “Full Stack Genome Engineering.”] Synthego, </ref> | In 1985, possibly the first major technique, for highly targeted genome editing, was the discovery of zinc finger nucleases (ZFN), which improved the effectiveness of gene targeting. ZFNs help “recognize” DNA, which makes it possible to design ZFNs for a variety of genomic targets varying throughout different cells and organisms. <ref> [https://www.synthego.com/learn/genome-engineering-history “Full Stack Genome Engineering.”] Synthego, </ref> ZFNs are fusion proteins composed of DNA-binding domains that recognize and bind to a specific set of three base pairs. They can be combined to recognize longer DNA sequences. For example, specificity to a nine-base-pair target sequence would require three ZFN domains fused in tandem. <ref> Fridovich-Keil, Judith L. [https://www.britannica.com/science/science "Gene Editing"] Encyclopædia Britannica, Inc., 20 Feb. 2020 </ref> Researchers then fused the DNA cleaving (cutting) enzyme Fok1 to the ZFN binding domain to essentially create “genomic scissors” which can cleave DNA at a specified location, creating a double stranded break (DSB) in the DNA. <ref> [https://en.wikipedia.org/wiki/Zinc_finger_nuclease “Zinc Finger Nuclease"] Wikipedia, Wikimedia Foundation, 29 Jan. 2020, </ref> A repair template is then introduced, so a fraction of the cells will undergo [https://en.wikipedia.org/wiki/Homologous_recombination homologous recombination], where the cell incorporates the gene of interest into its genome at the specific point. <ref> [https://www.synthego.com/learn/genome-engineering-history “Full Stack Genome Engineering.”] Synthego, </ref> | ||
===== Transcription Activator-like Effector Nucleases ===== | ===== Transcription Activator-like Effector Nucleases ===== | ||
| − | + | In 2007, the second generation of designer nucleases came in the form of transcription activator-like effector nucleases, or TALENs. These nucleases are able to recognize a single nucleotide rather than a trinucleotide motif like the ZFN. The TALENs benefit from ease of design, as effective TALENs can be designed and constructed in a few days, and can be multiplexed on the order of hundreds at a time. These nucleases work very similarly to ZFNs in that they are designed by fusing a DNA cutting domain of a nuclease to a TALE DNA recognition domain, which can be engineered to specifically recognize a unique sequence. These fusion proteins can then be used to form a targeted pair of “DNA scissors” that can be utilized to to perform targeted genome modifications such as sequence insertion, deletion, repair and replacement in living cells. TALENs offer 3 unique advantages compared to ZFNs. They have a higher DNA binding specificity, they have lower off-target effects, and they are easier to manufacture. <ref> [https://en.wikipedia.org/wiki/Genome_editing “Genome Editing.”] Wikipedia, Wikimedia Foundation, 25 Feb. 2020, </ref> | |
=== Advances in Gene Editing === | === Advances in Gene Editing === | ||
| + | |||
| + | |||
Revision as of 22:24, 14 March 2020
Genome Editing, also known as gene editing, is a group of technologies under genetic engineering that gives scientists the ability to change an organism’s DNA. This group of technologies enable genetic material to be added, removed, or altered at particular locations in the genome. Several methods of implementing foreign DNA into an organism's existing genome have been utilized in the past, however, the most recent is called Crispr-Cas9. Short for clustered regularly interspaced short palindromic repeats and CRISPR-associated protein 9, the Crispr-Cas9 system has generated a lot of excitement among the scientific community as it has been the fastest, cheapest, most accurate and efficient means of altering DNA compared to its predecessors. Genome editing is of great interest in the prevention and treatment of human diseases. Currently, most research on genome editing is done to understand diseases using cells and animal models. Scientists are still working to determine whether this approach is safe and effective for use in people. It is being explored in research on a wide variety of diseases, including single-gene disorders such as cystic fibrosis, hemophilia, and sickle cell disease. It also holds promise for the treatment and prevention of more complex diseases, such as cancer, heart disease, mental illness, and human immunodeficiency virus (HIV) infection. [1]
Contents
History
The idea of using gene editing to treat disease or alter traits is not a novel one and dates back to at least the 1950s and the discovery of the double-helix structure of DNA. In the mid 20th century, researchers realized that small changes in the DNA sequence could mean the difference between health and disease. This understanding led to the theory that by identifying “molecular mistakes” that cause genetic diseases we could fix those mistakes, enabling the prevention or reversal of these diseases. This concept was the underlying idea behind gene therapy through genome editing, and was seen as the future of molecular genetics. [2]
Early Forms of Gene Editing
Zinc Finger Nucleases
In 1985, possibly the first major technique, for highly targeted genome editing, was the discovery of zinc finger nucleases (ZFN), which improved the effectiveness of gene targeting. ZFNs help “recognize” DNA, which makes it possible to design ZFNs for a variety of genomic targets varying throughout different cells and organisms. [3] ZFNs are fusion proteins composed of DNA-binding domains that recognize and bind to a specific set of three base pairs. They can be combined to recognize longer DNA sequences. For example, specificity to a nine-base-pair target sequence would require three ZFN domains fused in tandem. [4] Researchers then fused the DNA cleaving (cutting) enzyme Fok1 to the ZFN binding domain to essentially create “genomic scissors” which can cleave DNA at a specified location, creating a double stranded break (DSB) in the DNA. [5] A repair template is then introduced, so a fraction of the cells will undergo homologous recombination, where the cell incorporates the gene of interest into its genome at the specific point. [6]
Transcription Activator-like Effector Nucleases
In 2007, the second generation of designer nucleases came in the form of transcription activator-like effector nucleases, or TALENs. These nucleases are able to recognize a single nucleotide rather than a trinucleotide motif like the ZFN. The TALENs benefit from ease of design, as effective TALENs can be designed and constructed in a few days, and can be multiplexed on the order of hundreds at a time. These nucleases work very similarly to ZFNs in that they are designed by fusing a DNA cutting domain of a nuclease to a TALE DNA recognition domain, which can be engineered to specifically recognize a unique sequence. These fusion proteins can then be used to form a targeted pair of “DNA scissors” that can be utilized to to perform targeted genome modifications such as sequence insertion, deletion, repair and replacement in living cells. TALENs offer 3 unique advantages compared to ZFNs. They have a higher DNA binding specificity, they have lower off-target effects, and they are easier to manufacture. [7]
Advances in Gene Editing
Use of Gene Editing
Ethical Issues
External Links
Further Reading
References
- ↑ "What Are Genome Editing and CRISPR-Cas9?" NIH.” U.S. National Library of Medicine, National Institutes of Health, 3 Mar. 2020.
- ↑ Fridovich-Keil, Judith L. "Gene Editing" Encyclopædia Britannica, Inc., 20 Feb. 2020
- ↑ “Full Stack Genome Engineering.” Synthego,
- ↑ Fridovich-Keil, Judith L. "Gene Editing" Encyclopædia Britannica, Inc., 20 Feb. 2020
- ↑ “Zinc Finger Nuclease" Wikipedia, Wikimedia Foundation, 29 Jan. 2020,
- ↑ “Full Stack Genome Engineering.” Synthego,
- ↑ “Genome Editing.” Wikipedia, Wikimedia Foundation, 25 Feb. 2020,